library("gamlss2")
## create family object with
## different link specifications
fam <- Kumaraswamy(a.link = shiftlog, b.link = "log")
## simulate data
set.seed(123)
n <- 1000
d <- data.frame("x" = runif(n, -pi, pi))
## true parameters
par <- data.frame(
"a" = exp(1.2 + sin(d$x)) + 1,
"b" = 1
)
## sample response
d$y <- fam$r(1, par)
## estimate model using the Kumaraswamy family
b <- gamlss2(y ~ s(x), data = d, family = fam)GAMLSS-RS iteration 1: Global Deviance = -1504.1261 eps = 0.674808
GAMLSS-RS iteration 2: Global Deviance = -1504.1299 eps = 0.000002 ## plot estimated effect
plot(b)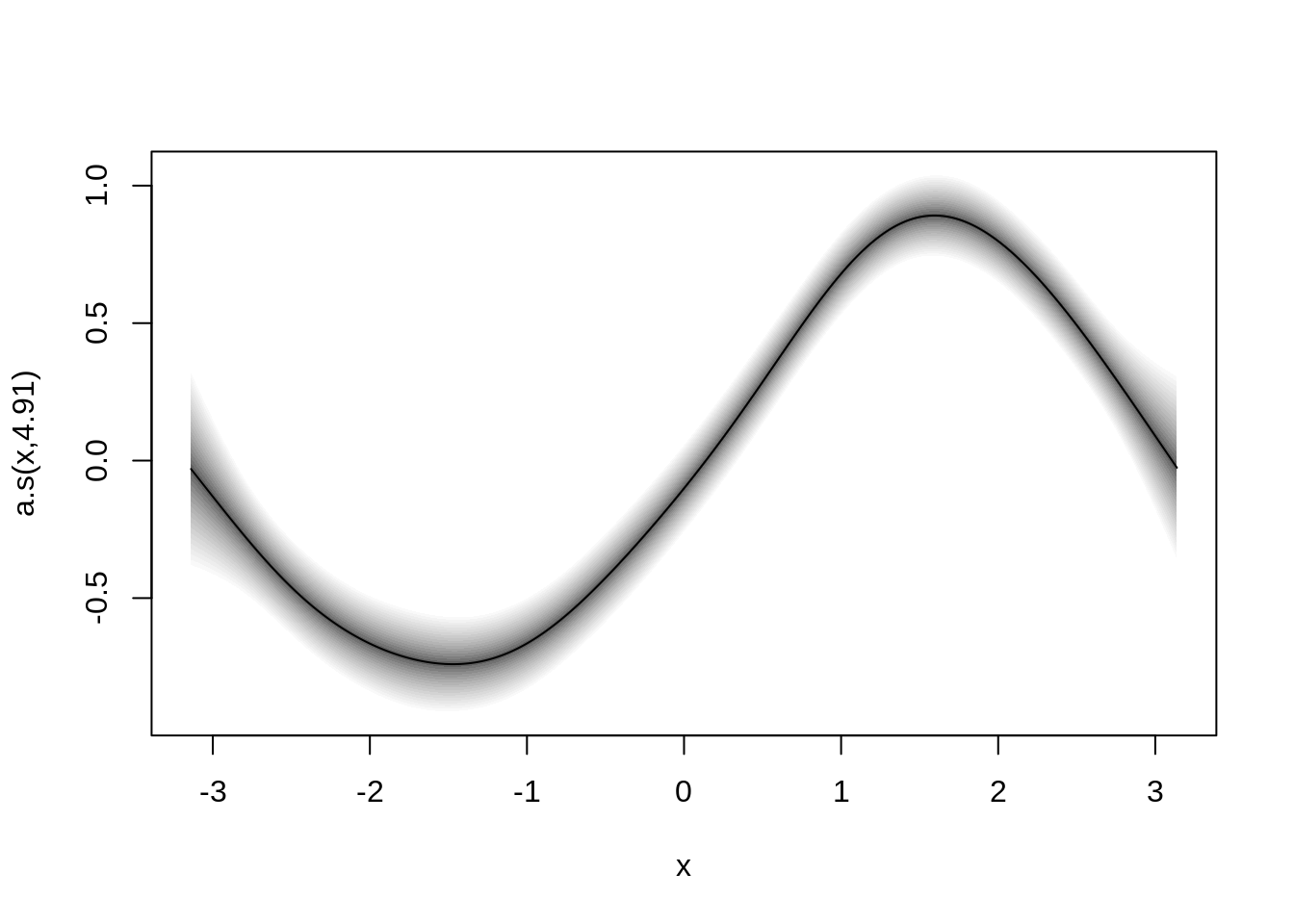
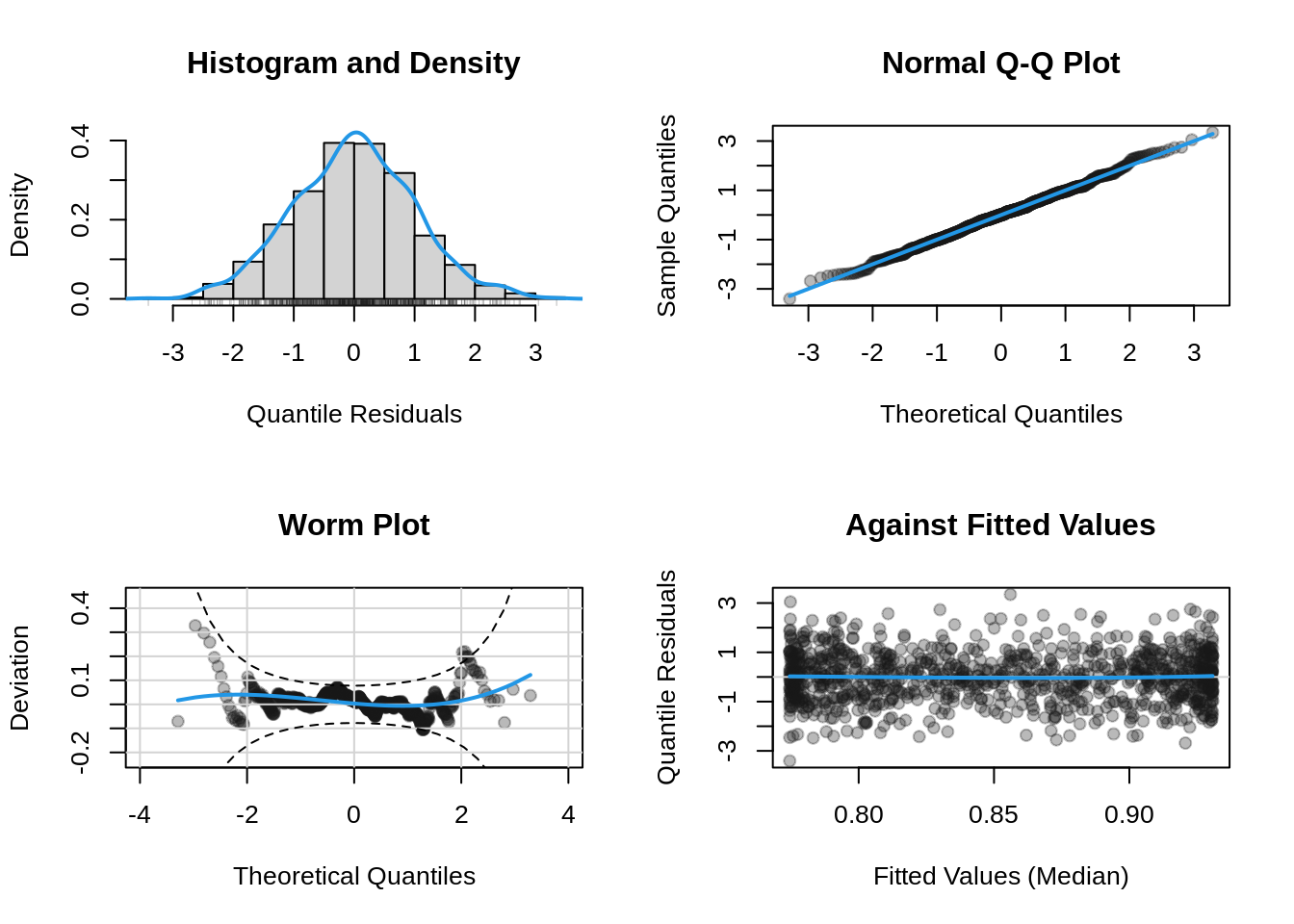
## plot residual diagnostics
plot(b, which = "resid")