library("gamlss2")
## visualization of softmax function from Wiemann et al. (2003, Figure 1)
x <- -200:200/50
plot(x, softplus(1)$linkinv(x), ylab = expression(softplus[a](x)),
type = "l", col = 2, lwd = 2)
grid()
lines(x, softplus(5)$linkinv(x), col = 3, lwd = 2)
lines(x, softplus(10)$linkinv(x), col = 4, lwd = 2)
lines(x, pmax(0, x), lty = 3, lwd = 2)
legend("topleft", c("a = 1", "a = 5", "a = 10", "linear spline"),
col = c(2, 3, 4, 1), lty = c(1, 1, 1, 3), lwd = 2, bty = "n")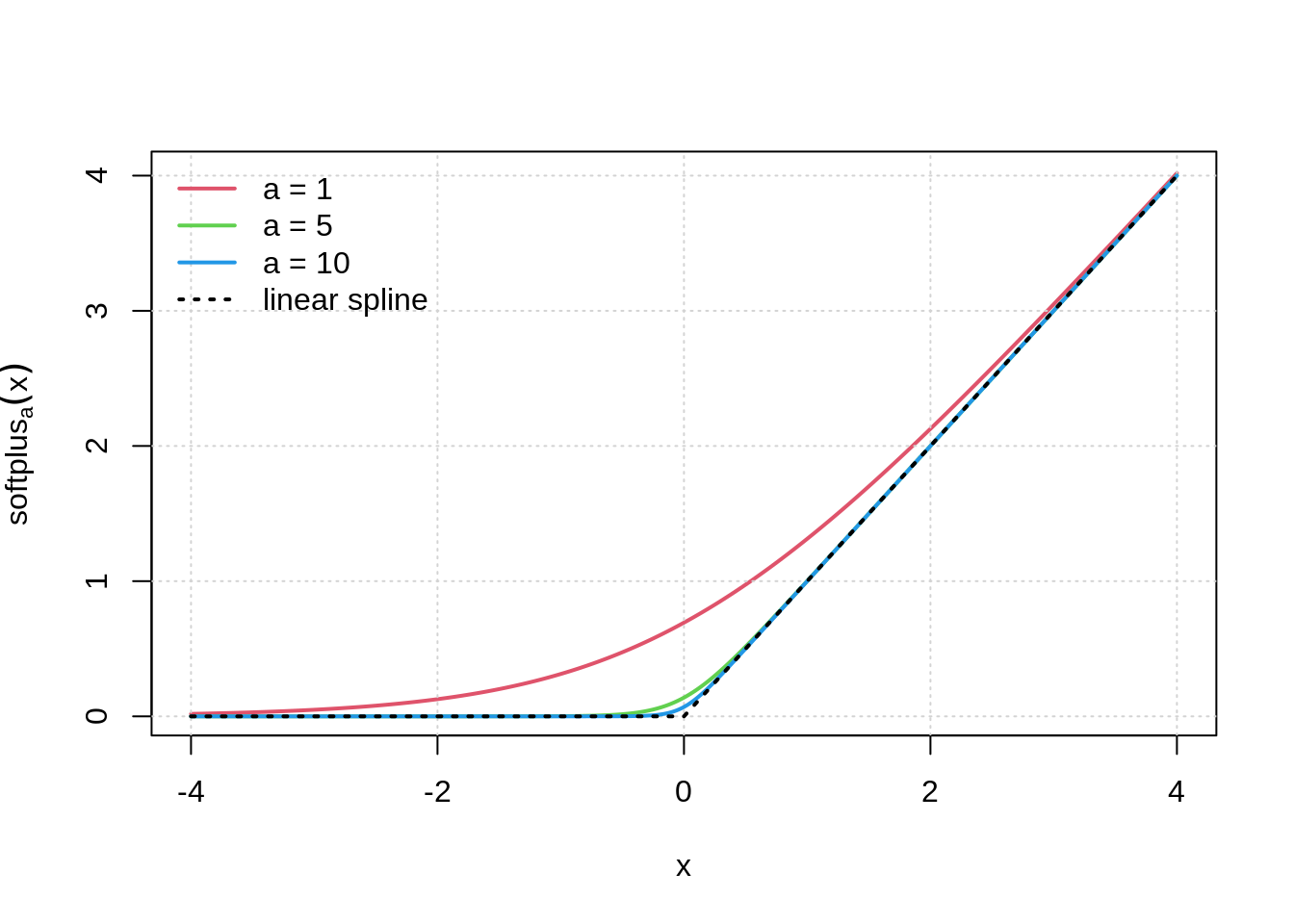
## Poisson regression example with different links
data("FIFA2018", package = "distributions3")
m_exp <- glm(goals ~ difference, data = FIFA2018, family = poisson(link = "log"))
m_splus <- glm(goals ~ difference, data = FIFA2018, family = poisson(link = softplus(1)))
AIC(m_exp, m_splus) df AIC
m_exp 2 359.3942
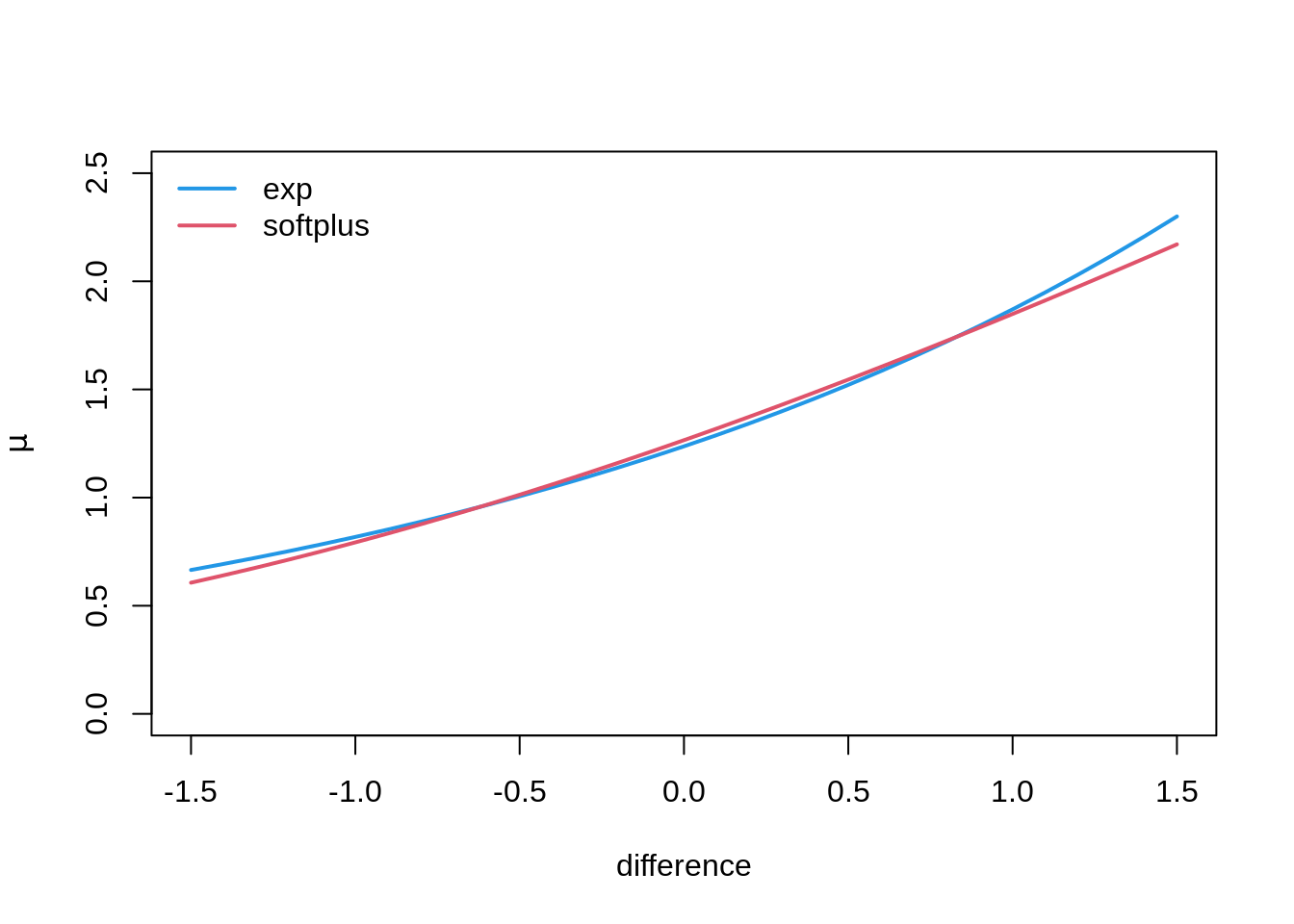
m_splus 2 359.3774## comparison of fitted effects
nd <- data.frame(difference = -15:15/10)
nd$mu_exp <- predict(m_exp, newdata = nd, type = "response")
nd$mu_splus <- predict(m_splus, newdata = nd, type = "response")
plot(mu_exp ~ difference, data = nd, ylab = expression(mu),
type = "l", col = 4, lwd = 2, ylim = c(0, 2.5))
lines(mu_splus ~ difference, data = nd, col = 2, lwd = 2)
legend("topleft", c("exp", "softplus"), col = c(4, 2), lwd = 2, lty = 1, bty = "n")