library("gamlss2")
## load head circumference data
data("dbhh", package = "gamlss.data")
## specify the model Formula
f <- head ~ pb(age) | pb(age) | pb(age) | pb(age)
## estimate model
b <- gamlss2(f, data = dbhh, family = BCT)GAMLSS-RS iteration 1: Global Deviance = 26202.5288 eps = 0.394214
GAMLSS-RS iteration 2: Global Deviance = 26201.9975 eps = 0.000020
GAMLSS-RS iteration 3: Global Deviance = 26201.9389 eps = 0.000002 ## visualize estimated effects
plot(b, which = "effects")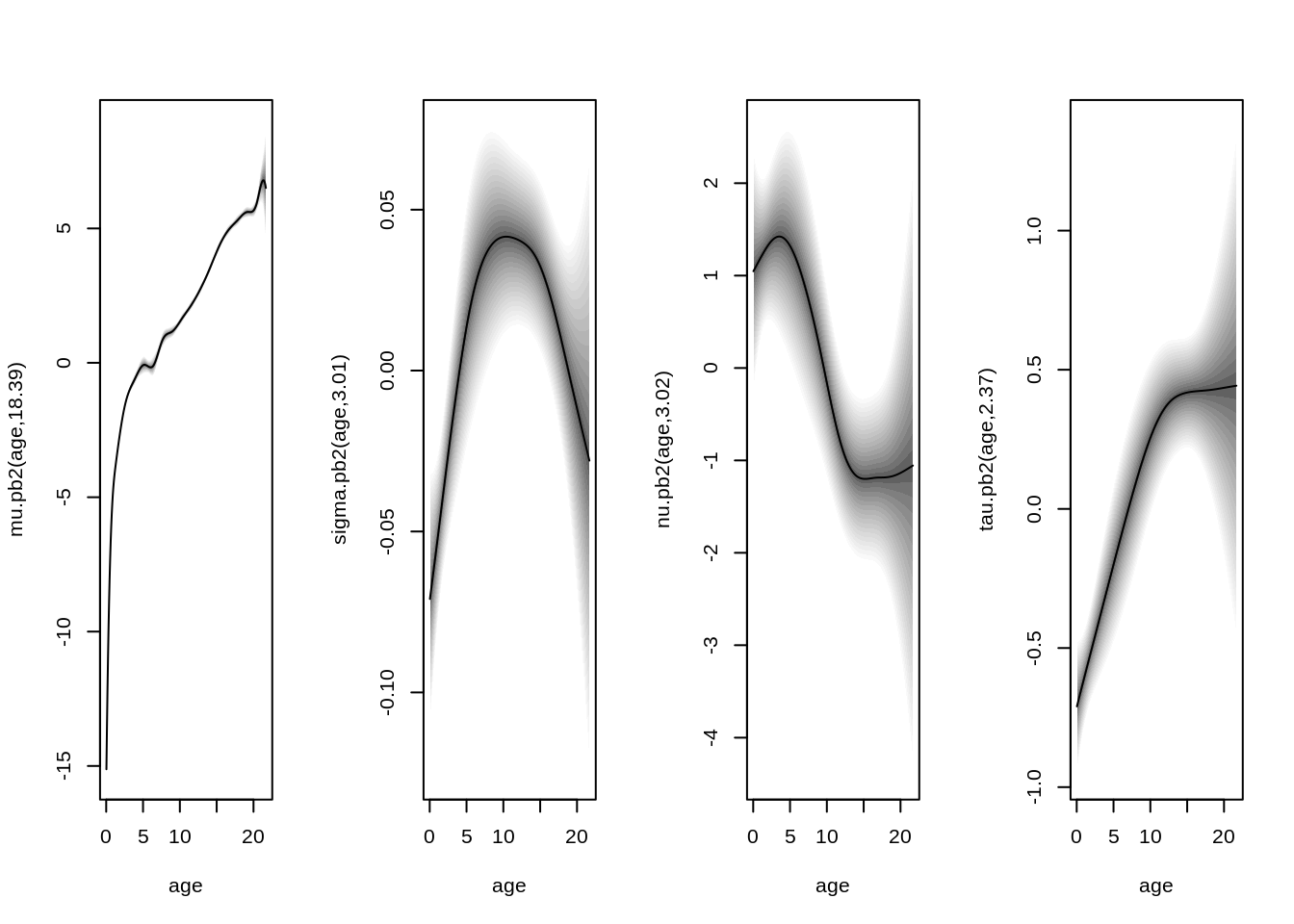
## plot diagnostics
plot(b, which = "resid")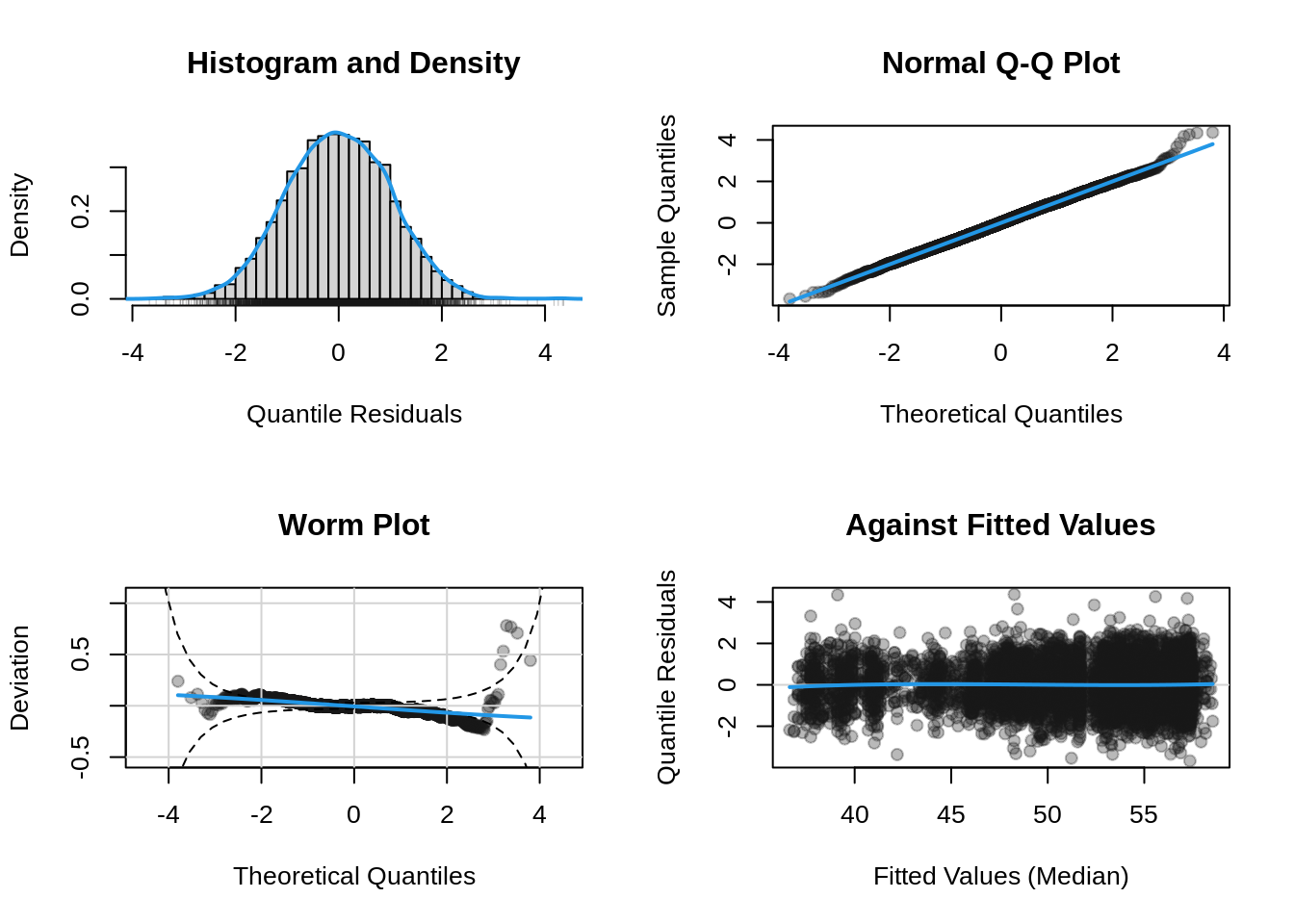
## predict quantiles
pq <- quantile(b, probs = c(0.05, 0.5, 0.95))
## plot
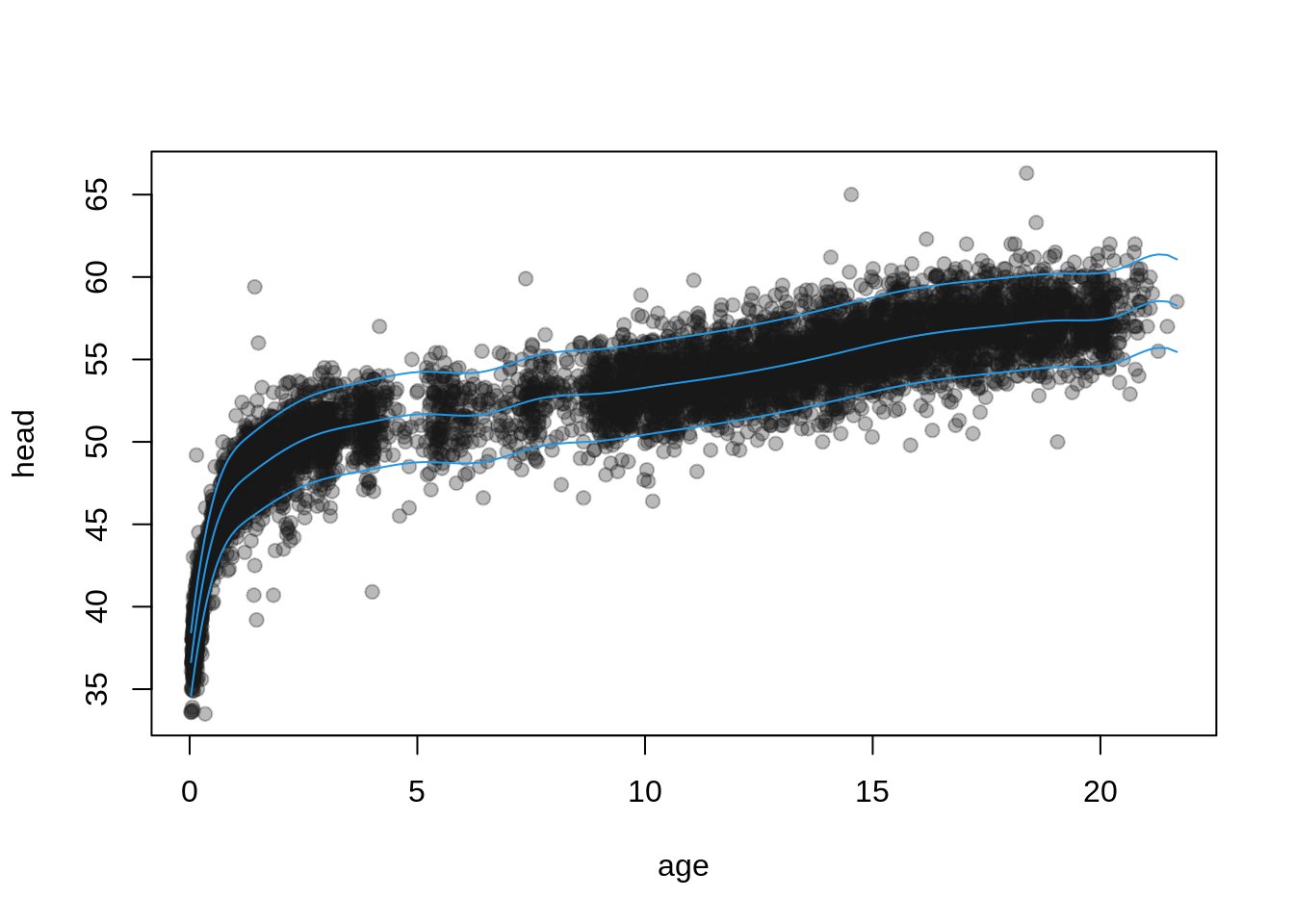
plot(head ~ age, data = dbhh, pch = 19,
col = rgb(0.1, 0.1, 0.1, alpha = 0.3))
matplot(dbhh$age, pq, type = "l",
lty = 1, col = 4, add = TRUE)